The novel coronavirus does not exist in isolation; it belongs to a larger virus family—Sarbecovirus. This family includes numerous members widely found in animals such as bats and pangolins, characterized by extremely high genetic diversity and frequent recombination capabilities. Multiple cross-species transmission events to humans (e.g., SARS-CoV and SARS-CoV-2) pose a long-term and severe threat to global public health security. However, most existing vaccines and antibody therapies target the receptor-binding domain (RBD) of the viral spike protein, which is highly prone to mutations, resulting in significantly reduced efficacy against emerging variants. Therefore, developing a new generation of antiviral agents that target conserved regions of the virus and possess broad-spectrum neutralizing activity has become an urgent core mission for both the scientific community and industry. The joint research team induced "super immunity" in camelids and successfully isolated 100 pan-sarbecovirus nanobodies (psNbs). These psNbs demonstrated potent neutralization efficacy against various sarbecoviruses, including SARS-CoV, SARS-CoV-2, and its variants (such as Omicron). Structural analysis of 13 psNbs revealed that they target five classes of conserved epitopes on the RBD, and their neutralizing potency is closely related to the distance from the epitope to the receptor-binding site (RBS). Additionally, the research team developed an inhalable bispecific psNb (PiN-31), offering a new direction for expanding anti-coronavirus intervention strategies.
Induction of "Super Immunity" and Antibody Screening
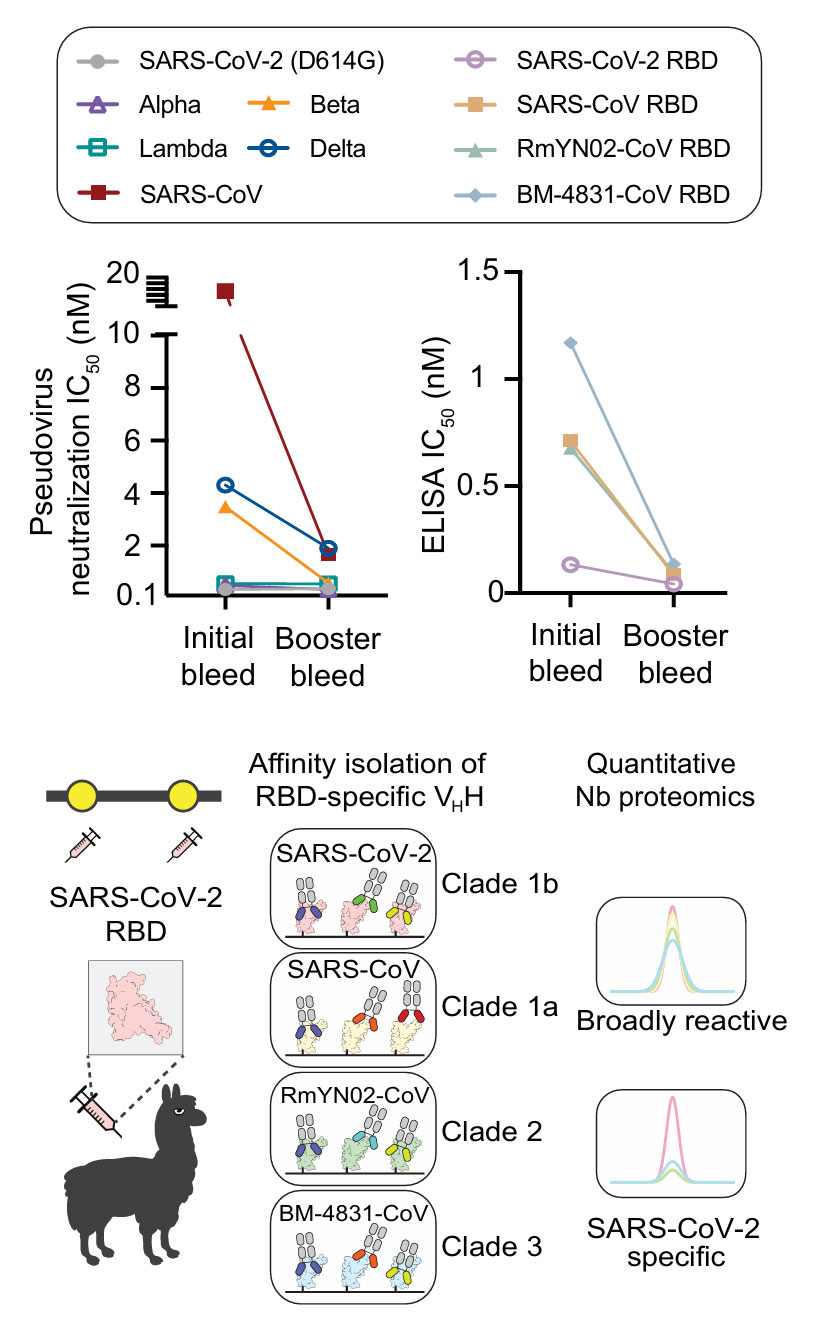
First, the researchers selected alpacas as immunization subjects and administered up to eight immunizations with SARS-CoV-2 RBD protein. The initial immunization used 0.2 mg of RBD mixed with complete Freund’s adjuvant, followed by seven booster immunizations, each with 0.1 mg of RBD. After multiple boosters, they observed a qualitative leap in the immune response of the alpacas: the single-domain antibodies (VHHs) in the serum not only exhibited higher affinity for the immunogen but also significantly expanded their binding and neutralizing breadth, showing enhanced activity against variants such as Beta and Delta, as well as the original SARS virus. This phenomenon is referred to as "Super Immunity."
Subsequently, the research team purified the plasma from the "super immune" alpacas and used RBD affinity capture technology to preliminarily screen out antibodies that bound to the RBD. Advanced integrated proteomics analysis, aided by the AugurLlama tool, successfully isolated and identified 100 high-affinity psNbs from numerous antibodies. These psNbs possess unique advantages: studies confirmed that all of them cross-react with the RBDs of sarbecoviruses from different clades, and 42% of the psNbs can simultaneously bind all four viral clades, demonstrating exceptional broad-spectrum activity. Seven psNbs not only exhibited strong cross-reactivity with the viral RBD but also showed no cross-reactivity with human whole-protein extracts, indicating high specificity.
Figure 1: Alpaca Immunization and psNb Discovery
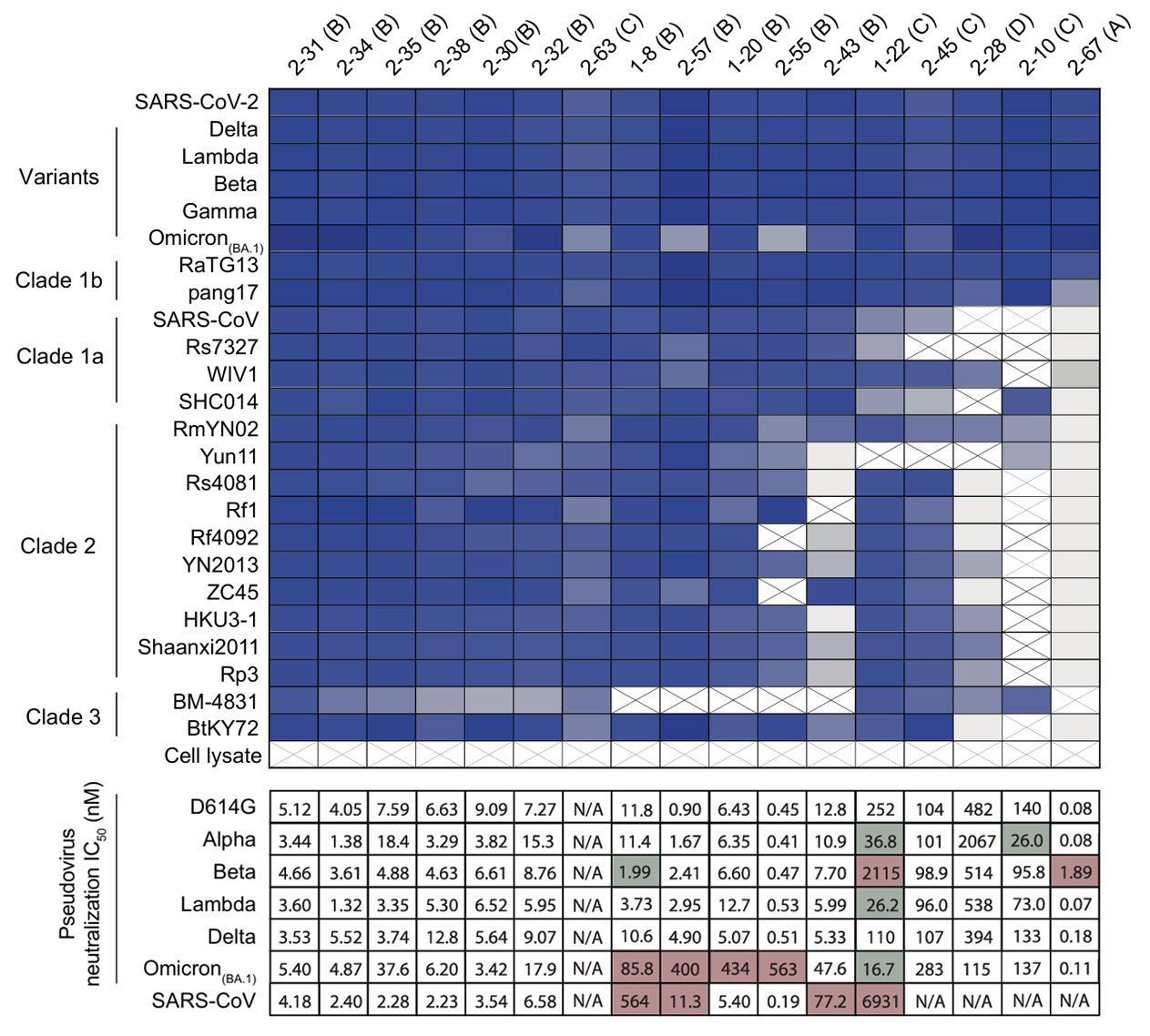
Further functional validation of these 100 psNbs revealed that most of them possess excellent neutralizing capacity. As shown in the figure below, representative psNbs exhibited neutralizing potency at nanomolar or even picomolar levels against various pseudoviruses, including Omicron, far surpassing many traditional antibodies.
Figure 2: Neutralizing Activity of Representative psNbs Against Different Variants and Sarbecoviruses
Structural Biology Reveals Epitope Characteristics and Mechanisms of psNbs
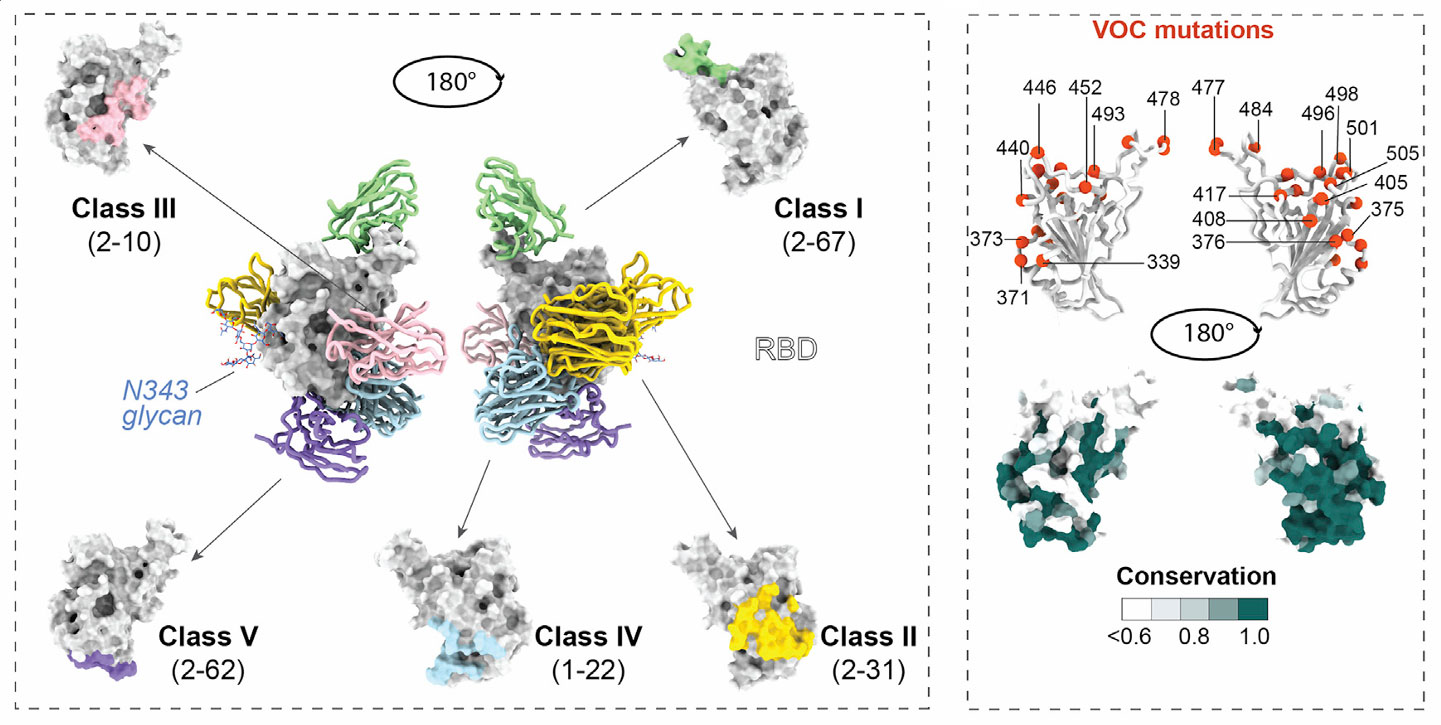
To gain deeper insights into the mechanisms of psNbs, the research team employed cryo-electron microscopy (cryo-EM) and X-ray crystallography for structural analysis. Using cryo-EM, they resolved the structures of 11 psNbs in complex with spike/RBD, while X-ray crystallography resolved the structures of 2 psNb-RBD complexes. Based on this structural information, the researchers classified the psNbs into five distinct epitope classes.
-
Class I: Represented by 2-67, targets the RBS (receptor-binding site), with its binding region located on the RBD ridge (aa472-490). These psNbs achieve potent viral neutralization by directly competing with ACE2 for binding sites, blocking virus-receptor interaction. They exhibit the highest neutralizing potency and account for 3% of all psNbs.
-
Class II: Includes 2-31, 2-57, etc., binds to non-RBS regions of the RBD and is further divided into subclasses II(A) and II(B). They share a hydrophobic core (aa377-386) and interfere with ACE2-RBD binding through steric hindrance. This class has the highest representation among all psNbs.
-
Class III: Such as 2-10, binds to rare non-RBS epitopes, partially overlapping with the Nb17 epitope but shifting towards a more conserved small region. These psNbs exert neutralization by destabilizing the spike structure, though their neutralizing potency is relatively weak.
-
Class IV: Represented by 1-22 and 2-45, targets RBD-up conformation-specific epitopes, binding to a conserved cavity (containing D427, F464, etc.). Although they do not directly compete with ACE2 binding, they neutralize the virus with moderate potency.
-
Class V: Like 2-62, binds to a conserved epitope inside the spike. While capable of binding RBD, it does not bind the pre-fusion spike and has almost no effective neutralizing activity, accounting for 38% of all psNbs.
Furthermore, the epitopes targeted by psNbs are highly conserved, comprising >75% conserved surface residues on the RBD, and avoid key variant mutation sites such as aa371, 373, and 375. Crucially, a strong negative correlation (Pearson r=0.95) was observed between neutralizing potency and the distance from the epitope to the RBS. This discovery provides key theoretical guidance for developing more effective antiviral molecules in the future.
Figure 3: Distribution of Binding Epitopes for the Five Classes of psNbs on SARS-CoV-2 RBD
Functional Validation of psNbs and Development of Bispecific psNb
Researchers assessed the binding capability of psNbs to 24 different RBDs using ELISA. Results showed that multiple psNbs exhibited strong binding activity, further confirming their broad binding spectrum. Simultaneously, Surface Plasmon Resonance (SPR) was used to determine the binding kinetics parameters of the psNbs. For instance, 2-31 showed a K_D <1 pM for Clade 1b RBD, indicating extremely high affinity. In neutralization assays, pseudovirus neutralization tests covering variants like Omicron, and plaque reduction neutralization tests (PRNT) against the Munich and Delta strains were conducted. Results demonstrated that psNbs possess strong neutralizing capacity against SARS-CoV-2 and its variants. For example, Class I psNbs (e.g., 2-67) had a median neutralizing potency of 1.4 ng/mL; Class II psNbs (e.g., 2-55) had a lowest potency of 7 ng/mL. Against SARS-CoV, Class II psNbs also showed high potency, and Class IV psNbs (e.g., 1-22) reached potencies in the single-digit µg/mL range.
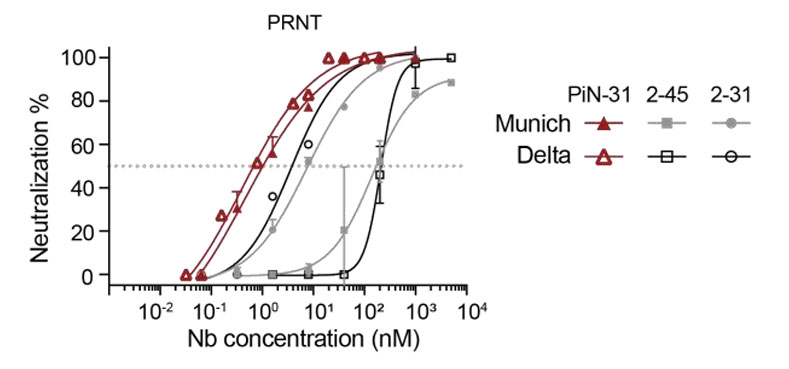
Finally, the research team fused 2-31 (Class II(B)) with 2-45 (Class IV) to create an inhalable bispecific nanobody, designated PiN-31. This bispecific molecule covers two different classes of conserved epitopes, offering unique advantages. PRNT results showed that PiN-31 achieved a neutralizing potency as high as 0.4 nM, an order of magnitude improvement over the monomers, demonstrating enhanced neutralization. Moreover, psNbs themselves are highly stable, tolerate aerosolization without loss of activity, making PiN-31 suitable for inhalation administration, directly targeting the respiratory tract. This provides a new effective strategy for preventing and treating respiratory infectious diseases.
Figure 4: Schematic Diagram of the Efficacy of Bispecific Nanobody PiN-31
The research conducted by the University of Pittsburgh team represents not only a technological breakthrough but also an innovation in strategic thinking. It suggests that instead of passively chasing evolving viruses, we should proactively utilize advanced technologies like nanobodies to target the virus's Achilles' heel. These findings have broad application prospects and significant market value. These psNbs, especially in bispecific or multivalent formats, can provide effective design strategies for broad-spectrum vaccines or therapeutics for prophylactic or therapeutic use.