Research jointly published by the Laboratoire de Recherche en Sciences Végétales (Toulouse, France) and the Light Imaging Center in the Cell Press journal Plant Communications provides a novel method to overcome the technical challenges in plant protein research. This study successfully validated the broad applicability of the ALFA tag/ALFA nanobody system in plants, enabling a full range of applications from subcellular localization and protein interaction analysis to super-resolution imaging and targeted degradation. This technology specifically breaks through the research barriers for hard-to-tag proteins like the Arabidopsis IRT1 iron transporter, providing a versatile, highly compatible innovative tool for plant protein functional analysis, thereby reshaping the paradigm of plant biology research.
Construction of a Plant Expression System for ALFA Nanobody-Fluorescent Protein Fusions
The research team first constructed an ALFA nanobody expression system suitable for plants. They genetically engineered an Arabidopsis codon-optimized ALFA nanobody (NB) fused to fluorescent proteins such as mCitrine or mScarlet, driven by the UBI10 promoter. The UBI10 promoter was chosen for its ability to provide mild constitutive expression, avoiding the gene silencing issues common with the CaMV 35S promoter.
Following Agrobacterium-mediated transformation of Arabidopsis, single-insertion homozygous T3 lines were selected. Phenotypic analysis revealed no significant differences in root length or growth rate between transgenic and wild-type plants, demonstrating that the ALFA NB-fluorescent protein fusions are non-toxic. Confocal microscopy observation showed fluorescent signals widely distributed in tissues such as root tips, root hairs, and mesophyll cells, with the proteins primarily localized to the cytoplasm and nucleus, laying the foundation for subsequent binding to target proteins.
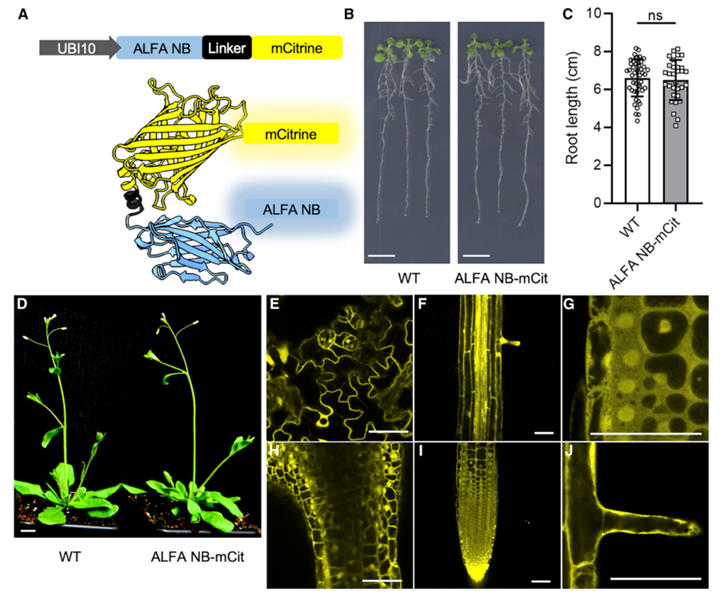
Figure 1: Expression of ALFA Nanobody-Fluorescent Protein Fusions in Plants
Validation of Subcellular Localization Detection Capability
To validate the ALFA system's ability to mark different subcellular structures, researchers fused the ALFA tag to known marker proteins and co-expressed them with ALFA NB-fluorescent proteins.
Results showed that when the plasma membrane protein Lti6b-ALFA was co-expressed with ALFA NB-mCitrine, fluorescence shifted from the cytoplasm/nucleus to the plasma membrane, co-localizing with FM4-64, proving precise plasma membrane protein labeling. After wortmannin treatment, the protein accumulated in late endosomes, consistent with the known endocytic pathway. When the microtubule-associated protein (MAP4) fused with an ALFA tag was co-expressed with ALFA NB-mScarlet, typical microtubule filamentous structures were observed, which were disrupted by oryzalin, validating the effective labeling of cytoskeletal proteins. For the nucleus and mitochondria, an ALFA-tagged protein carrying a nuclear localization signal (NLS) successfully guided ALFA NB-mScarlet into the nucleus. The mitochondrial outer membrane protein OM64-ALFA completely co-localized with ALFA NB-mScarlet. However, as nanobodies cannot penetrate membrane structures, the system could not detect proteins in the endoplasmic reticulum lumen or mitochondrial inner membrane.
These results indicate that the ALFA nanobody system can efficiently label proteins in various subcellular structures including the cytoplasm, nucleus, plasma membrane, and mitochondria (outer membrane).
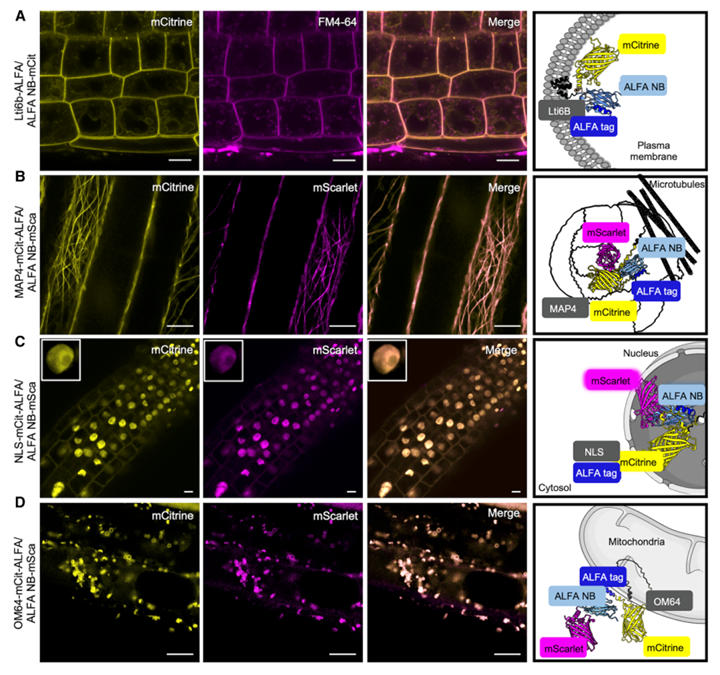
Figure 2: Detection of ALFA-Tagged Proteins via Fluorescent Protein-Fused ALFA Nanobodies
Breakthrough in Studying the Hard-to-Tag Protein IRT1: Nanobodies Unlock Functional Analysis
Arabidopsis IRT1 is a key transporter for iron uptake. Its complex multi-transmembrane domain topology means that N- or C-terminal tagging often disrupts its function. Traditional fluorescent protein insertion is only effective in specific extracellular loops, frequently leading to loss of function. The research team utilized the ALFA nanobody system to achieve functional tagging and analysis of IRT1.
-
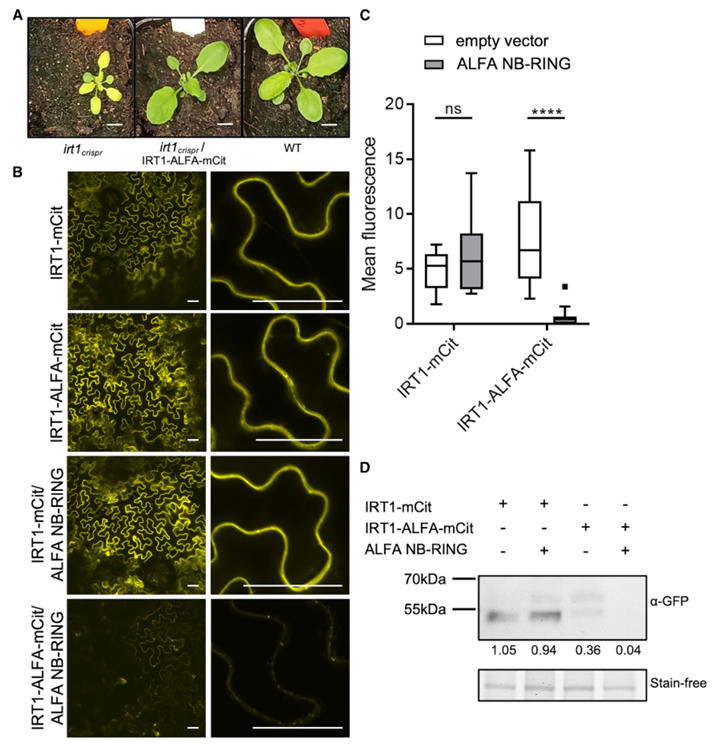
Tag Insertion Optimization: Researchers inserted the ALFA tag into four potential sites of IRT1. They found that fusion proteins with insertions in extracellular loop 1 (P41-C42) and intracellular loop 3 (T148-S149) could fully complement the iron deficiency chlorosis in the irt1 mutant. The IRT1-ALFA construct with the intracellular loop 3 insertion retained 100% functionality, whereas inserting mCitrine at the same site caused protein retention in the endoplasmic reticulum, highlighting the ALFA tag's compatibility with complex topologies. The intracellular loop insertion was more conducive to ALFA NB binding and performed better than fluorescent protein fusions at the same position.
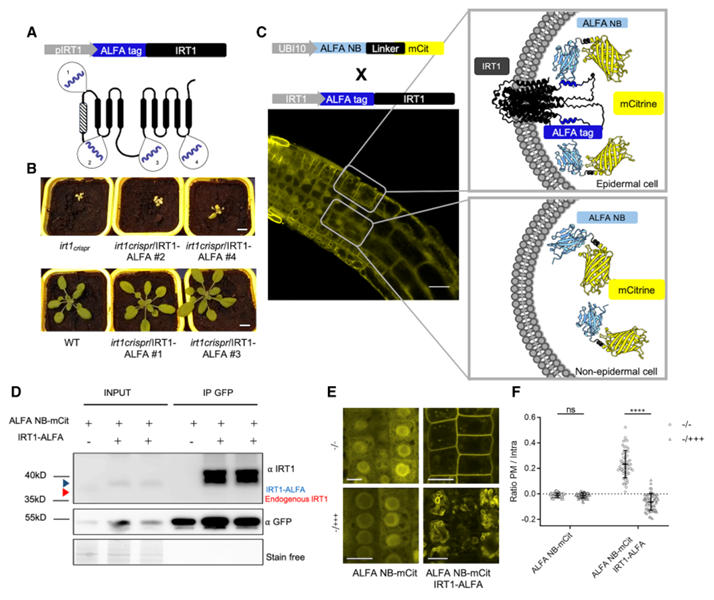
Figure 3: Proof of Concept for ALFA Technology Applied to IRT1 Imaging and Co-Immunoprecipitation
-
Localization and Dynamic Regulation: IRT1-ALFA localized to the plasma membrane in root epidermal cells, consistent with the expression pattern of endogenous IRT1. Under conditions of excess non-iron metals, IRT1-ALFA was translocated to the vacuolar membrane via endocytosis, a process that could be clearly tracked using ALFA NB-fluorescent proteins, aligning with known metal regulation mechanisms.
-
Interaction Validation: Using the ALFA system to construct a Ternary Fluorescence Complementation (TriFC) assay, the interaction between IRT1 and its known partners AHA2 (H⁺-ATPase) and FRO2 (ferric chelate reductase) was confirmed. This approach resolves the false-positive issues caused by signal peptide cleavage in traditional Bimolecular Fluorescence Complementation (BiFC).
-
Targeted Degradation: Fusing the ALFA NB to the RING domain of an E3 ubiquitin ligase specifically induced the degradation of IRT1-ALFA without affecting untagged IRT1, demonstrating the system's utility for precise protein function regulation.
Figure 4: Targeted Degradation of ALFA-Tagged IRT1 Protein
Super-Resolution Imaging: Nanobodies Enable Single-Molecule Tracking
Traditional super-resolution microscopy techniques rely on direct fusion of the target protein to photo-convertible fluorescent proteins (e.g., mEos3.2). However, the large size of these tags (>25 kDa) can interfere with protein function, especially for multi-transmembrane proteins, often causing failure.
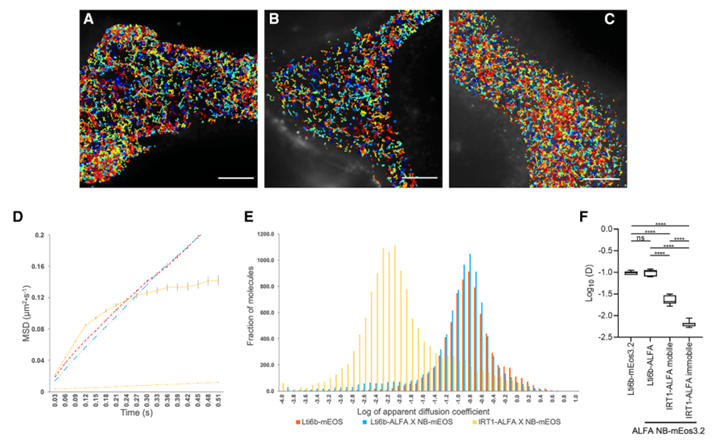
To enable super-resolution tracking of plant proteins, the research team innovatively designed a "ALFA tag - nanobody - photo-convertible fluorescent protein (mEos3.2)" ternary system. The study employed an indirect labeling strategy: the ALFA tag is inserted into the protein of interest (POI), and a genetically encoded ALFA nanobody carrying mEos3.2 achieves specific binding. Dynamics are then tracked using sptPALM (single-particle tracking photoactivation localization microscopy). To validate the system's reliability, the plasma membrane protein Lti6b was used as a reference, comparing the effects of direct fusion (Lti6b-mEos3.2) versus indirect labeling (Lti6b-ALFA + ALFA NB-mEos3.2).
Results showed no significant difference in the diffusion coefficient of Lti6b between the two labeling methods. Both groups captured over 7400 valid trajectories with comparable trajectory density, fully demonstrating that mediation by the ALFA nanobody does not interfere with normal protein dynamics and validating the technical effectiveness of the ternary system.
Figure 5: Super-Resolution Imaging of Lti6b-ALFA and IRT1-ALFA via sptPALM
As a key iron uptake transporter in Arabidopsis, IRT1's complex multi-transmembrane structure and functional sensitivity mean that direct fusion with traditional fluorescent proteins often leads to its retention in the ER or loss of transport activity, making normal study impossible. The ALFA nanobody, however, has a small molecular volume, and the ALFA tag is only 17 amino acids long. The nanobody's specificity is extremely high, binding only to the ALFA tag, and its impact on target protein function is almost negligible. Furthermore, the ALFA nanobody system uses a "single transgenic event" strategy: once the ALFA-tagged fusion protein is constructed, multiple applications can be achieved using pre-existing ALFA nanobody lines, significantly enhancing experimental efficiency.
From a scientific perspective, this study represents the first quantification of single-molecule dynamics for a complex transmembrane protein in living plants. It not only provides a feasible path for functional studies of hard-to-tag proteins like IRT1 but also establishes a new technical paradigm for plant membrane protein research, advancing plant biology to the single-molecule level. Moreover, this research not only validates the ALFA system's revolutionary potential for plant protein studies but also provides an efficient tool for analyzing important crop traits, offering molecular instruments for deciphering mechanisms of nutrient transport and stress response, thereby promoting precision design breeding.

Wuhan Nano Body Life Science and Technology Co. Ltd. (NBLST) is a nanobody industry platform established under the initiative of the Wuhan Industrial Innovation and Development Research Institute. Its headquarters is located in the main building of the Wuhan Industrial Innovation and Development Research Institute in the East Lake High-tech Development Zone, Wuhan. It boasts a 1400 m² independent laboratory in the Precision Medicine Industrial Base of Wuhan Biolake. Additionally, NBLST has established alpaca experimental and transfer bases in Zuoling, Wuhan, and Tuanfeng, Huanggang, both compliant with laboratory animal standards. These bases currently house over 600 alpacas, providing "zero-immunization-background" guaranteed alpaca immunization services for research institutions and antibody drug development companies.
NBLST focuses on the development, engineering, and application of nanobodies, and is dedicated to building an integrated public experimental service platform for production, education, and research. It possesses a full-chain technology platform encompassing antigen preparation (peptides, proteins, and RNA), antibody discovery and engineering, through to biological function validation/screening. The RNA antigens include RNA structurally and sequentially optimized for alpacas. Antibody discovery and engineering services employ multiple technological routes, including phage display, RNA, and mammalian cell display. Through cross-complementation of multiple platforms, it provides flexible antibody discovery and engineering services for pharmaceutical companies and research institutes, facilitating the development of drug reagents.
In addition to its natural nanobody library, NBLST also offers an off-the-shelf immunized library to help clients quickly screen for antibody molecules that meet their needs.
If you require our services, please feel free to contact us via email: marketingdept@nanobodylife.com